The ABEL trap (“anti-brownian electrokinetc trap”) has been invented and realized by A. E. Cohen and W. E. Moerner in Stanford
(see websites of AEC in Harvard and WEM in Stanford).
It’s purpose is to immobilize single molecules, proteins or nanoparticles in solution by active canceling their three dimensional Brownian motion. The nano-object is monitored by fluorescence imaging. The position of the nano-object is determined with respect to a target position and used for a feedback voltage applied to four electrodes. The electric field or the induced electroosmotic flux pushes the nano-object back and compensates the Brownian motion.
A microfluidic device constricts the diffusion of the nano-object to the x and y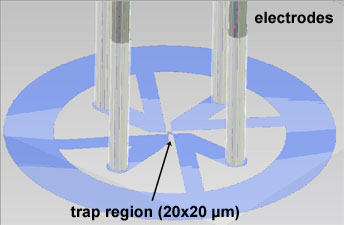
dimensions. The chamber is made of transparent PDMS. Four 20 µm deep channels
with the electrodes are arranged perpendicular to confine the nano-object within the trap region with only 1 µm thickness.
Widefield illumination of the trap region and a fast sensitive CCD camera to detect the position of the fluorescent nano-object was sufficient to immobilize a single DNA molecule, a tobacco mosaic virus (300 nm length), a liposome (100 nm radius) or even a 10 nm polystyrene bead. For trapping smaller objects like single fluorophores or soluble proteins, faster versions of the ABEL trap have been developed.
 Application to a single FoF1-ATP synthase
Application to a single FoF1-ATP synthase
(monitoring conformations of a single membrane transporter)
In collaboration with AEC and WEM we modify the ABEL trap to immobilize a single liposome with one embedded membrane proteine. Therefore the FoF1-ATP synthase is labeled with two fluorophores to monitor the internal rotary motion by confocal single-molecule FRET. and is transported to the target position in the ABEL trap.
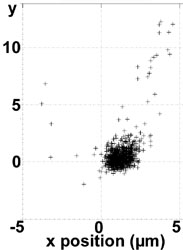
Actually we can trap 100 nm liposomes (see image on the left: the liposome is confined for seconds before the fluorescence intensity falls below a trapping threshold due to photobleaching; then the liposome diffuses away in the upper right direction) and are implementing the combined FRET @ ABELtrap experiment.
Movie of an Atto680-labeled liposome trapped for several seconds in the ABEL trap (Flash file):
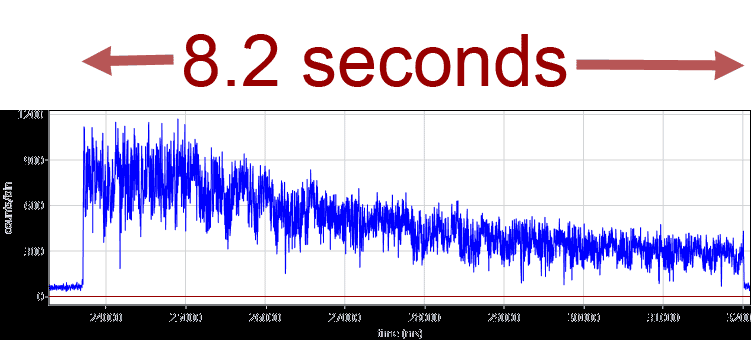
The faster versions of the ABEL trap now use a confocal laser pattern and single-photon-based feedback from APDs. Localization analysis and electrode feedback are implemented in a FPGA chip. Fluorescent lipids, which might interfere with the single-molecule FRET measurements of proteins, are not required anymore because the feedback utilizes the photons of the single FRET fluorophors. We have built a modified ‘hardware ABELtrap’ and have captured 20-nm fluorescent beads for more than 8 seconds in contrast to their mean diffusion time of 10 ms within the PDMS microfluidics. A time trajectory of an ABELtrapped fluorescent 20-nm bead is shown below.
Now we study the conformational dynamics of single FoF1-ATP synthases at different ATP concentrations for the first time.